- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - GGAGAT
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - GAGATC
- -tgattgt
- - - - - - - - - - - - - - - - - - - -ggtggtg
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - cgtggtg
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - tgtgttg In case of intronic elements, one can click the sequence to see the corresponding data: a WebLogo for a cluster that contains the putative SRE, a corresponding PWM and score value that was calculated based on the PWM.
U12 INTRONS In this page one can browse and search for U12 introns predicted in all plant species except C. reinhardtii which is devoid of U12 splicing machinery. The most prominent features of U12 introns i.e. highly conserved sequence at donor site, terminal dinucleotide at acceptor site and branch site are marked with different colors as in example:
#21
gene: GRMZM2G154267, transcript: GRMZM2G154267_T01
Orthologs: AT5G22110 GLYMA16G34470 LOC_Os05g06840 LOC_Os08g36330 SELMODRAFT_236997 SELMODRAFT_111965 SELMODRAFT_236823 Vv17s0000g04830
CAATTGACTTATCTAATGCCatatcctaaaaaaacagtgttattgttagcacttatggctagttactatt ... gatttcgagattatcgagaaagtatcactactttccttaacatagaacacAAAATCACGTCAGGTTTTTT The provided orthologs represent orthologous genes with U12 intron found. Upon clicking the gene name, the U12 intron data in the gene is displayed. microRNA GENES One can choose a microRNA of interest from one of the following sources of miRNA gene structure: ERISdb predictions, miRNAs annotated in Ensembl Plants 15 or paper by Mica et al..
In case of Ensembl miRNAs, the user is redirected to a corresponding record in Splice site selection page.
Upon selecting a microRNA from ERISdb the following will be displayed (see example below):
- pre-miRNA sequence (yellow)
- EST sequence (green)
- genomic sequence (blue).
Predicted intron is in lower case and is truncated to 50 bases at both ends.microRNA: gma-MIR394gIn case of miRNAs from Mica et al. the alignment of miRNA splice site and deep sequencing reads are presented (see below). Here, reads that support the splice sites are chopped into two parts: one aligned to exon sequence at 5'ss and another - at 3'ss. For vvi-MIR162 and vvi-MIR168 there are multiple splice sites predicted but we present them separately as their arrangement in different splice forms is unknown.
EST name: AW099182.1
ACAGAGTTTCTTGGCATTCTGTCCACCTCCACTTCTTGGCCCTATCTACGTACTCGGAGGTGGATATACTGCCAATAGAG
CACGTGGGTTTAACAAAGGGTTGCTTACAGAGTTTCTTGGCATTCTGTCCACCTCCACTTCTTGGCCCTATCTACGTACTCGGAGGTGGATATACTGCCAATAGAGCTGTGTTGGCTTCTCTTTGTCAAGCCTCCGTGATACTATAAATCTCAGTTAAATACATAAACACTTTTTTCCCATTTTTCTTTTCTGGCTATCAATTATATGAG GATCCCACCGGTCGTTTGCAAAAAAATGCTAAGAAATCGGATAAAGAGTAGAACGCCTCGAGTCGCTTGCCGCTTGCTTCTGGTGTTAGAAAGAAACCCTATAATCTTGTTTCCCTCCATTCTTATGAAACCCTATATTTCCCTTAGAACTTGGGCCTTTCCATGTTTCTTTAATTCTACTAGTCTAGCGATTGTTTGATGTTTCTAGTTTATTTCTTAAAATAAATTCTTGACGTTGTGT
CACGTGGGTTTAACAAAGGGTTGCTTACAGAGTTTCTTGGCATTCTGTCCACCTCCACTTCTTGGCCCTATCTACGTACTCGGAGGTGGATATACTGCCAATAGAGCTGTGTTGGCTTCTCTTTGTCAAGCCTCCGTGATACTATAAATCTCAGTTAAATACATAAACACTTTTTTCCCATTTTTCTTTTCTGGCTATCAATTATATGAGgtaaagcaacctccttcctttttctttccttttgttgttttgtttttgtg ... ctagatagcacgatactgtgtgcttttcccctcattttttgattttgcagGATCCCACCGGTCGTTTGCAAAAAAATGCTAAGAAATCGGATAAAGAGTAGAACGCCTCGAGTCGCTTGCCGCTTGCTTCTGGTGTTAGAAAGAAACCCTATAATCTTGTTTCCCTCCATTCTTATGAAACCCTATATTTCCCTTAGAACTTGGGCCTTTCCATGTTTCTTTAATTCTACTAGTCTAGCGATTGTTTGATGTTTCTAGTTTATTTCTTAAAATAAATTCTTGACGTTGTGT
ATGC exonic sequence ATGC 5' end of a read ATGC intronic sequence ATGC 3' end of a read vvi-MIR394B chr18 - 1385724 1385362 CTCTCTCGCTCTTCCACTCTAGAGCATCAAGGTGAAAACCCCA ........ CTTGTGTTGCAGGGGTTTCATCAACTCCTCCTCTTTGCCTCTT CTAGAGCATCAAG GGGTTTCATCAACTCCT 1 TCTTCCACTCTAGAGCATCAAG GGGTTTCATC 1 GAGCATCAAG GGGTTTCATCAACTCCTCCT 2 TCCACTCTAGAGCATCAAG GGGTTTCATCA 1 CATCAAG GGGTTTCATCAACTCCTCCTCTT 2 TTCCACTCTAGAGCATCAAG GGGTTTCATC 5 AGAGCATCAAG GGGTTTCATCAACTCCTCC 3WEBLOGOS
This page provides WebLogos for 53'ss, 33'ss and branch sites of U2 and U12 introns for analyzed plant species. also, the corresponding PWMs are available. DOWNLOAD
Different types of data and tools are available for download here.EXAMPLE
Click "Browse" in the main menu and select Zea mays. Then type "GRMZM2G134756" in the "Transcript selection" page (Input gene name or its part field).
There are two transcripts for this gene: GRMZM2G134756_T01 and GRMZM2G134756_T02: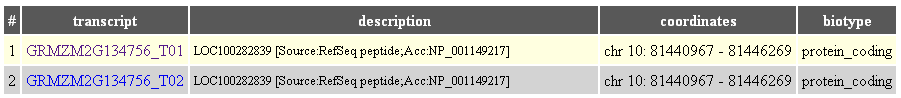
Click GRMZM2G134756_T01 and then choose 3' splice site of intron 4, as shown in the figure:
Note that intron 3 is marked with
, which means that it is more than 1000 bases long and was shortened for display purposes.
Five data tabs will appear: General, Orthologs, EST, PTT, BS, UA-tract, and cis-regulatory elements:

By clicking the tabs, one can access the corresponding information.
Let's look into "General" tab. It says that it is U12 intron and there is a link provided to U12 page. In the U12 page we can find out that both splice forms have U12-type intron and that this is a U12-type intron in Oryza sativa as well.
atgc highly conserved terminal intronic nucleotides at 5' or 3' end.
atgc branch site.#1If we go back to "Splice site data" page, in the orthologs tab we can see alignments of Zea mays splice site with two homologs in Oryza sativa. By clicking the transcript ids we can access the corresponding "splice site data'.
gene: GRMZM2G134756, transcript: GRMZM2G134756_T02
Orthologs: LOC_Os08g23110 LOC_Os08g05490
ACCAATTATTCTTTCACATCgtatcctgaatacttattttgatcttgttctctttttaacctttaaatat ... ttgtttgtactttgcagagcatgtatttttttgcttaacctgaaatgcagATATGCTCCCTGGTTTTAAA
#2
gene: GRMZM2G134756, transcript: GRMZM2G134756_T01
Orthologs: LOC_Os08g23110 LOC_Os08g05490
ACCAATTATTCTTTCACATCgtatcctgaatacttattttgatcttgttctctttttaacctttaaatat ... ttgtttgtactttgcagagcatgtatttttttgcttaacctgaaatgcagATATGCTCCCTGGTTTTAAA
In EST tag there is an experimental support for this splice site in a form of EST sequences. The two homologous splice sites in O. sativa are supported by both EST sequences and RNA-Seq reads.
All three homologs lack PPT tract but they have quite distinct UA-tracts (PTT, BS, UA-tract tab).
Finally, in the cis-regulatory elements tab we can discover that the homologs possess the same three putative regulatory sequences in exonic sequences.